
By Helen Hill for MGHPCC
Researchers in Greg Fournier's Geobiology Lab at the Massachusetts Institute of Technology (MIT), are using the MGHPCC facility in work seeking to calibrate the ancient history of life on Earth using the ultra youthful tool of genomic analysis.

Bubble of oxygen emerging from a cyanobacterial mat growing in the lab - image courtesy: Tanja Bosak, MIT
Scientists know that atmospheric oxygen irreversibly accumulated on Earth around ~2.3 billion years ago, at a time known as the Great Oxidation Event, or GOE. Prior to that time all life was microbial, and most, if not all, environments were anoxic (that is, contained no oxygen). Oxygen was first produced some time before the GOE through the evolution of a group of photosynthetic bacteria known as Cyanobacteria. Releasing oxygen as a by-product of splitting water in order to acquire electrons to be energized by light, this process led to dramatic changes in both the biological and geochemical processes on a planetary scale. Eventually, the continued accumulation of oxygen led to an oxidized surface, atmosphere, and ocean that persist to this day.
Besides shedding light on a fundamental change in Earth's climate, it is hoped that understanding the GOE will help scientists gain insight into the rise of eukaryotes (cellular organisms like us, in which the genetic material is DNA in the form of chromosomes contained within a distinct nucleus.). Eukaryotes require oxygen to produce sterols, an important part of their cell membranes. Furthermore, eukaryotes also contain mitochondria, organelles descended from ancient bacteria that use oxygen to generate energy using aerobic respiration.
There are currently two schools of thought regarding how oxygen levels rose: The first proposes a small initial rise at the time of the GOE, with levels low but stable until increasing again around 600 million years ago, approaching modern levels. The second posits a more oscillatory rise with a greater increase immediately following the GOE, and then a subsequent crash, with levels only increasing again 600 million years ago.
While geologists have been able to establish increasingly precise dates for the onset of the GOE through geochemical analyses, the ability to detect transient variations in oxygen levels following the GOE are less readily detected in the rock record. However, in the past couple of decades, it would be fair to say, science has experienced a GGE or Great Genomics Event through which biologists, armed with the ability to sequence genes increasingly rapidly, now find themselves hard at work sequencing everything they can lay their hands on. And it turns out genomics may hold the answer to how oxygen continued to accumulate,
Greg Fournier, an assistant professor of geobiology in the Department of Earth, Atmospheric and Planetary Sciences at MIT, is an expert in molecular phylogenetics, discovering the evolutionary histories of genes and genomes within microbial lineages across geological timescales.
A particular current interest is detecting events in the evolution of microbial metabolisms that likely align with global changes in Earth’s biogeochemical cycles, including oxygen.
Molecular oxygen (O2) readily changes to an extremely reactive “free radical” form with an unpaired electron called superoxide, a chemical highly damaging to many biological systems. Many organisms are protected against superoxides by Superoxide dismutase enzymes that convert superoxide to hydrogen peroxide, the first step in detoxifying this compound. It is present in most extant bacteria (ie ones that are alive today) but is assumed to have originally appeared in response to the increasingly oxygen rich environment of the GOE.
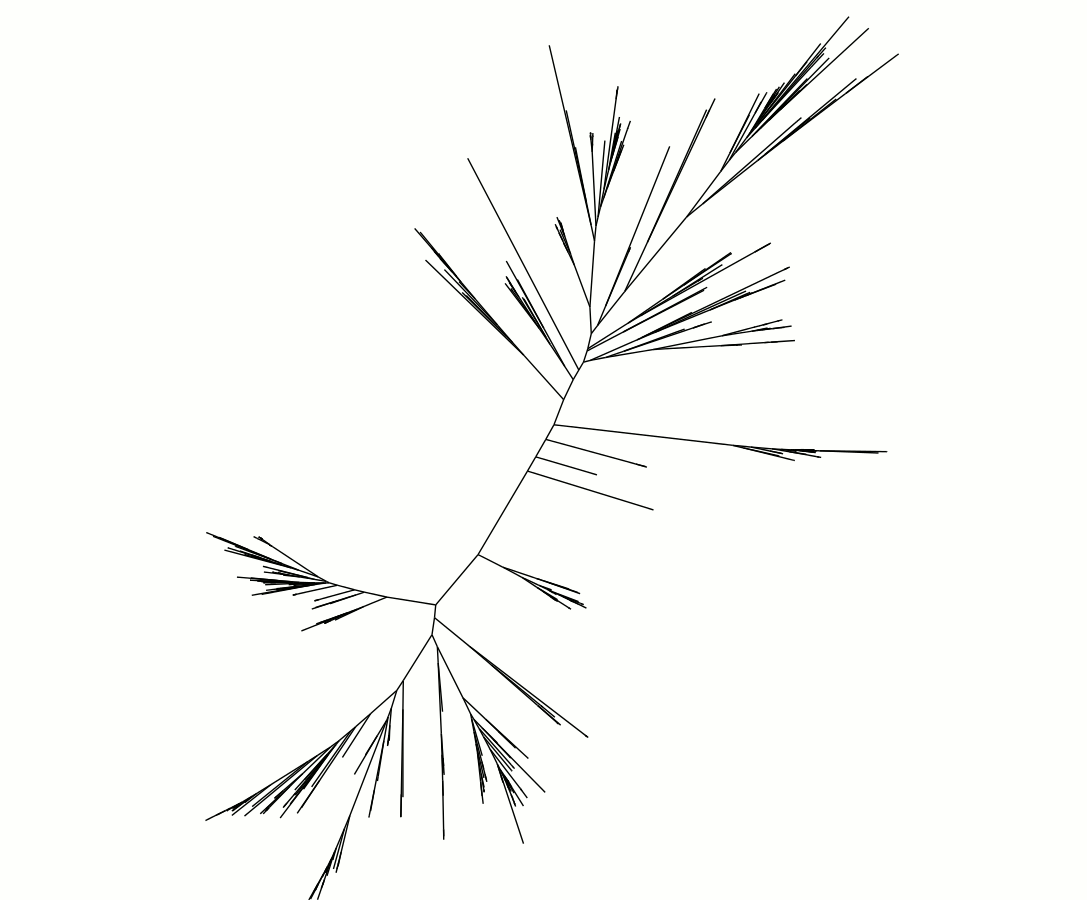 A simple tree image of only a portion of the Superoxide Dismutase genes within one part of the Tree of Life (Archaea), containing 500 species. The tree has no root because it remains to be determined where the ancestor branch should go. The MGHPCC Cluster allows the Fournier Group to make trees containing over 8000 species, across all of Bacteria as well as Archaea, generating vast amounts of tree data - image courtesy: Greg Fournier
A simple tree image of only a portion of the Superoxide Dismutase genes within one part of the Tree of Life (Archaea), containing 500 species. The tree has no root because it remains to be determined where the ancestor branch should go. The MGHPCC Cluster allows the Fournier Group to make trees containing over 8000 species, across all of Bacteria as well as Archaea, generating vast amounts of tree data - image courtesy: Greg Fournier
Fournier is an expert in a process called Horizontal Gene Transfer, or HGT. HGT is the exchange of genetic material between cellular organisms other than by regular "vertical" transmission of DNA from parent to offspring. He believes HGT evidence of oxygen-related genes like Superoxide dismutase will enable him to distinguish between a steady and a fluctuating build-up.
"If oxygen rose and remained steady we should see many such transfer events associated with Superoxide dismutase," Fournier explains. "If it rose and then fell back we would expect to see transfer events followed by the disappearance of the gene in different lineages, since the need to protect against oxygen would have ceased."
Because genetic data from old extinct lineages is not available, members of Fournier's Lab use gene sequences sampled across modern organisms, building evolutionary trees (phylogenies) to explore how they relate to one another. By comparing these gene trees to the best guesses of how the microbial organisms are related transfer events may be detected, and their relative timing inferred.
 Abigail Caron - image credit: Helen Hill
Abigail Caron - image credit: Helen Hill
Abigail Caron, a postgraduate researcher in the Fournier Group, uses a computer cluster housed at the Massachusetts Green High Performance Computing Center (MGHPCC) to run genetic analyses on different bacteria looking for instances of horizontal gene transfer, and mapping these events across many lineages.
For only a small number of gene sequences Caron can use (Rapid ANalysis of Gene Family Evolution using Reconciliation DTL or Ranger DTL) run on her laptop. But seeking to compare and integrate gene histories across upwards of 8000 bacterial species, incorporating complex models of uncertainty within individual tree analyses, as she is attempting to do, is too intensive for any single computer. Having the MGHPCC cluster to work on allows her to run multiple analyses simultaneously across dozens of processors, making such high-resolution investigations into the history of these genes possible.
Special thanks to Dr Jo Wolfe, as well as Abigail Caron and Prof Greg Fournier for their help in preparing this article.
Greg Fournier is the Cecil and Ida Green Assistant of Geobiology in the Department of Earth, Atmospheric and Planetary Sciences (EAPS) at the Massachusetts Institute of Technology (MIT.) His research integrates phylogenetics and horizontal gene transfer (HGT) with studies of microbial evolution, geochemistry, and planetary history. Specific areas of his research include: HGT- and genome-based calibration of molecular clock models of microbial evolution; ancestral reconstruction of ancient proteins and metabolisms; the biogeochemical impact of HGT and microbial metabolism evolution; and the role of partial HGT in the complex ancestry of organismal lineages.