In the face of the COVID 19 pandemic, a Massachusetts biotech startup turns to the MGHPCC for HPC resources in its hunt for existing FDA-approved drugs that might be a therapeutic candidate against the novel coronavirus.
Reporting by Helen Hill for MGHPCC
Joy Alamgir is a Massachusetts-based serial entrepreneur pursuing ideas in biotechnology with a computational focus. Alamgir Research, the startup he currently leads, brings together expertise in computer science, virology, clinical medicine, epidemiology, physics, and mathematics. When SARS-CoV-2 hit, Alamgir, recognizing the usefulness of the modeling tool his company has been developing, immediately repurposed, pivoting effort to simulating FDA-approved active compounds with the goal of rapidly identifying existing ones able to interrupt SARS-CoV-2 proteins.
Viruses replicate by entering cells and commandeering their reproductive machinery. SARS-CoV-2 makes multiple proteins to propagate. If these proteins can be safely and effectively targeted, then viral activity and replication can be inhibited, resulting in reduced disease severity.
Alamgir and his team are currently working through a list of over 1200 drug active compounds using Commonwealth Computational Cloud for Data-Driven Biology (C3DDB) high-performance computing resources housed at the MGHPCC to virtually stimulate which already-FDA-approved drugs can perform such viral protein interruption. “Since vaccine development, manufacturing and distribution for COVID-19 will be a lengthy process, identifying existing FDA approved compounds that can be repurposed to interrupt SARS-CoV-2 proteins is a priority“, says Alamgir.
So far Alamgir and his collaborators have found multiple FDA-approved drugs that can interrupt SARS-Co-V-2 proteins in-silico. "We have completed simulations for 5 out of the 25 SARS-Co-V-2 proteins and have discovered multiple FDA-approved drugs that can interrupt these proteins at the molecular level," Alamgir says.
Alamgir is confident in his team’s approach. ”Our modeling gives a quantitative measurement of protein interruption effectiveness since our quasi-quantum simulations focus on both molecular energy and molecular interaction characteristics,” explains Alamgir. “In layman’s terms: we are using super-computing to find the proverbial needles in a drug haystack of over a thousand drugs. Furthermore, since our search is focused on existing drugs, the drugs’ side effects are already known, and industry already has the manufacturing capability. Thus, the problem is reduced to, ‘Can an existing drug be effective?’ – which is exactly the answer we are searching for.”
With a well-defined to-do list - headed by super-computing based simulations of the remaining proteins - next steps for the most promising include pharmacological analysis, retrospective epidemiological studies, in- vitro petri-dish based viral testing, and finally, in-vivo small animal-based viral testing.
Alamgir and his team plan to make the analyzed results from their simulations (and later in-vitro and in-vivo validation results) publicly available. “Although we are a for-profit company, this work transcends that as this is a wide societal need,” he says.
Alamgir Research is looking for external grant funding/donations to offset significant costs related to in-vitro and in-vivo experimental tests.
To find out more about this work contact Alamgir
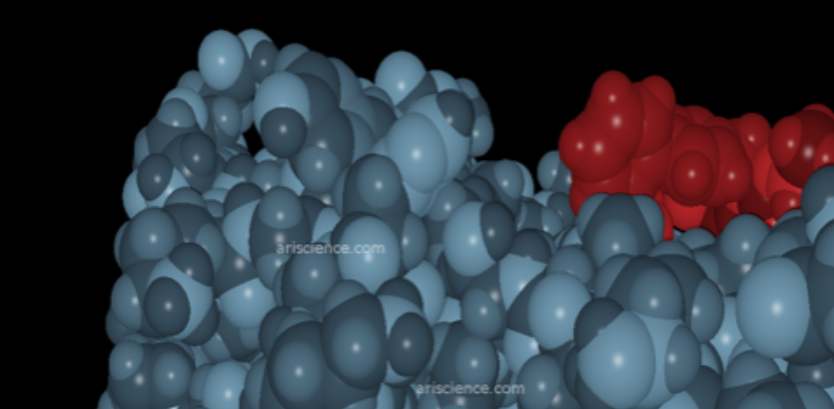
Story Image: Example protein interaction visualization - courtesy Alamgir Research. Using high-performance computing to perform picometer-scale molecular modeling, Alamgir Research simulates how key proteins COVID 19 uses to propagate behave in the presence of existing FDA-approved therapies.
Joy Alamgir is an entrepreneur based in Massachusetts currently pursuing ideas in biotechnology with a computational focus under the umbrella Alamgir Research Inc. He has previously started a public health software company whose software is still being used widely in the United States and Australia to manage 90+ communicable diseases, including but not limited to SARS-CoV-2 contact tracking and outbreak management (and previously for management of H1N1, Ebola, and other outbreaks). He grew the company into an epidemiology software market leader and sold it to Xerox in 2014. He completed his undergraduate in Computer Science and Economics at Cornell University and dropped out of Cornell’s Master of Engineering program with a semester left to go. 16 years later he went back to college to study biochemistry, genetics, oncology, immunology, virology, and quantum physics by dropping into specific courses at Harvard and WPI. He is also an advisor to multiple startups in construction, transportation, and health sectors in Massachusetts. He spends his non-work time with his wife and three children and practices martial arts to release steam.
The Commonwealth Computational Cloud for Data-Driven Biology (C3DDB) is a computer system dedicated to supporting research that connects life science research with emerging, innovative big data analytics. Use of the C3DDB is open to groups conducting life sciences research or advanced development at academic institutions, research institutes, and companies in Massachusetts. This includes both science and engineering researchers and developers of software tools and toolchains
Alamgir Research is a cross-disciplinary collaboration that includes leading experts in computer science, virology, clinical medicine, epidemiology, and mathematics. Joy Alamgir, the Principal Investigator, has worked in public health and big-data/big-computation for over 15 years and was the primary author of the system used by nearly 1/3rd of the United States for outbreak management including SARS-CoV-2 hotspots like New York City and Massachusetts. Dr. Ruhul Abid is part of the team for his clinical and experimental expertise. Rosa Ergas and Nicholas Hill are part of the team as senior epidemiologists. Dr. Mohsan Saeed and Dr. Masanao Yajima are part of the team as virology and statistics experts. Alamgir Research is based in Wayland, MA. Its mission is “Better and faster drug design and drug re-positioning through blending extreme science and engineering concepts.”
This work has also benefited from NSF/XSEDE, and the University of Maine System computing resources.