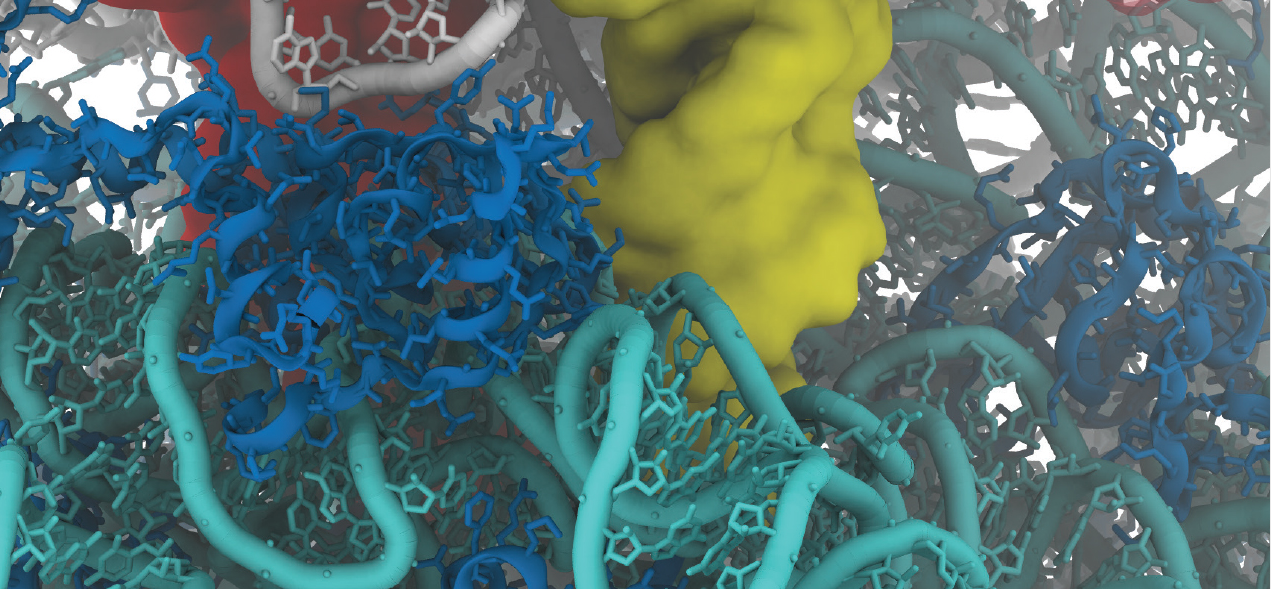
Paul Whitford is an assistant professor of physics at Northeastern University. He uses high performance computing to study the dynamics of biological systems to understand the physical principles that govern the dynamics of cells.
A particular interest is bacterial ribosomes. "With the ribosome we are trying to understand the dynamics of a very complex molecular system," says Whitford. In this video, Whitford lifts the lid on the interior landscape of the ribsosome, and his research seeking a nanoscale biomechanical understanding of how it works.
Modeling Molecular Machines - video credit: Helen Hill
"You can think of these systems as tiny motors similar to a car engine except scaled down by billions in size. What we are trying to understand, is how these little motors travel between all the different places in the cell and how they carry out all of their various functions when they get there."
To do this Whitford and his team run elaborate molecular simulations on Northeastern University's Discovery Cluster housed at the MGHPCC in Holyoke, MA. "If you think of a car engine, you know where the gas pedal is and what it does - well, put simply, our research is asking where is the gas pedal in these little machines?"
The research that Whitford's Group is pursuing has many applications ranging from health to nano-technology. "The nano-technology applications we have in mind are the design of molecular scale machines to carry out highly specific functions within the body. Our work provides the rules for how those machines work and blueprints for designing them. On the health side perhaps you would like to suppress replication of a cancer cell or kill some bacteria. To do that you have to know what are the key features that control each biological system."
Whitford recalls how when he first started trying to model the complexities of the ribosome back when he was a graduate student at the University of California, San Diego his colleagues would look at him like he was a little bit crazy, saying that what he was attempting was impossible. Now though, with the rapid increase in computing power and the facilities accessible to him through Northeastern and MGHPCC, he is able to run such models routinely making it practical to propose many ideas, and test them with rapid turn around times that really allow the science to move quickly, something that just ten years ago, he says, would have been unthinkable.
"Now that we have so many resources, and we have so many models that we can apply to study these systems, we can start to tackle the many conformational changes and functions of the system," says Whitford. In their work his team has so far touched on just two or three major motions, but he says there are dozens that remain unexplored and he is looking forward to what the future holds. "It's going to take many years to really go through each of these steps and understand what is going on through each of them, but the potential pay off is enormous," he says.