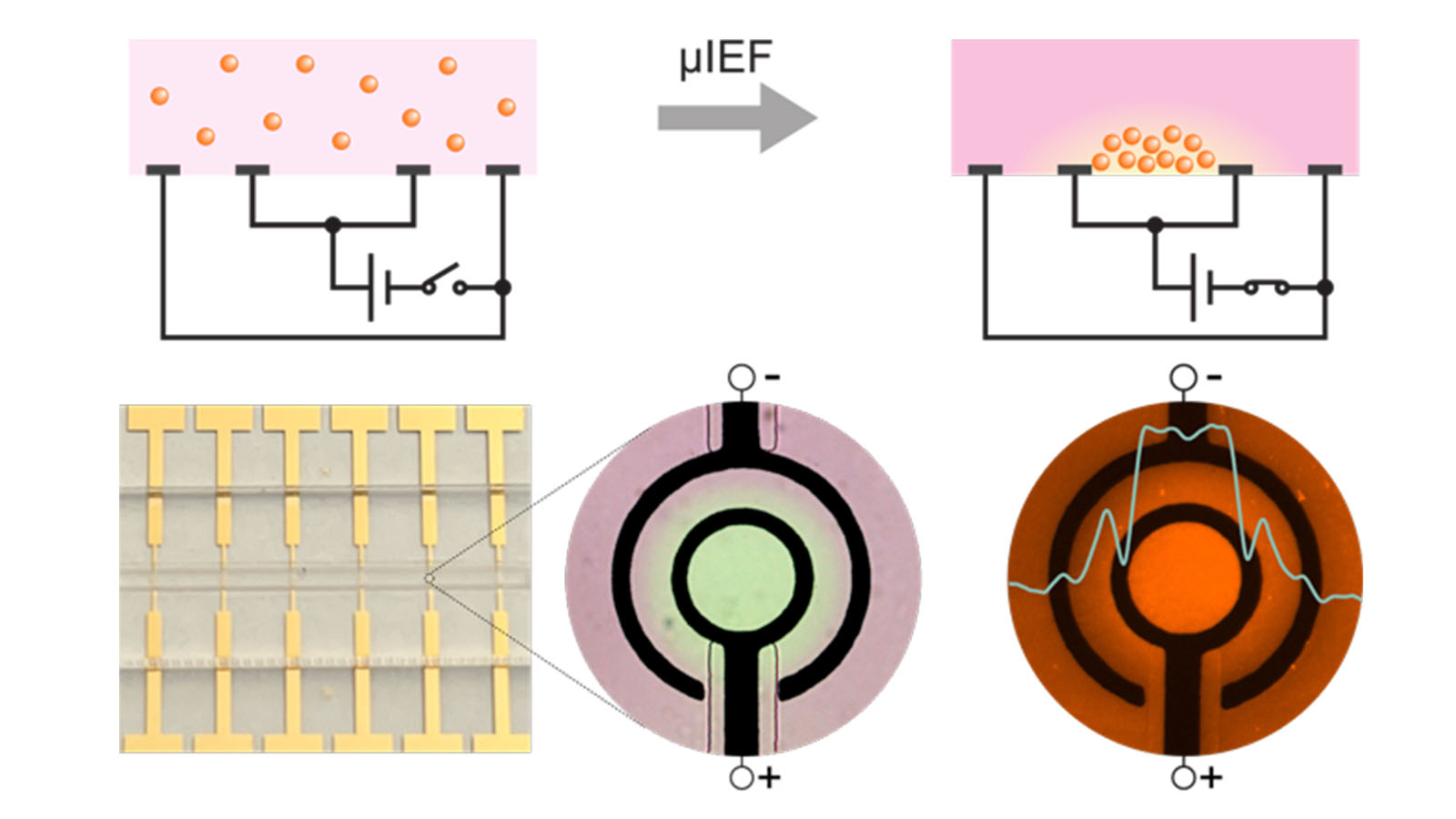
Many studies in biology and medicine – including those involved in clinical diagnostics, many therapeutics, and large-scale studies of proteins – rely on detecting protein concentrations in samples. Such studies often depend on detecting minute protein concentrations and are thus constrained by the protein-detection limit of the sensors used. Faced with this limitation, researchers are forced to employ more-expensive detectors or more-complicated assays. In response to this challenge, a research team led by Jinglei Ping – an assistant professor in the UMass Amherst Mechanical and Industrial Engineering (MIE) Department who is also affiliated with the Institute for Applied Life Sciences – has developed a groundbreaking method for concentrating proteins in such samples, amplifying the detection limit in their protein analysis, and measuring these protein concentrations less expensively and more effectively.
Ping and his teams’ revolutionary method employs an inexpensive isoelectric focusing technique, which can detect proteins at concentrations four times lower, and therefore four times more effective, than is currently possible.
The Ping Nano/Bio Interfaces and Applications Lab uses computers housed at the MGHPCC.